Applying Atomistic Modeling to Predict NLGN3 Isoform Binding to Neurexin 1- Beta
Why this study matters!
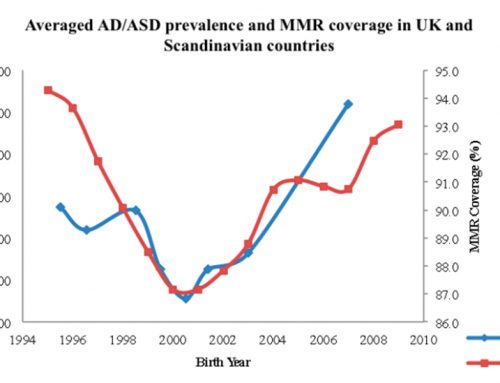
Autism spectrum disorders have been associated with over 300 genetic mutations. As a polygenic disease, ASDs require at least one additional insult in addition to the underlying genetic susceptibility in order for manifestation of the disease phenotype (Caldwell, 2010). For example, Bradley et. al. (2010) have identified large de novo deletions in NRXN1 (neurexin 1) exons associated with ADHD probands. Notably, many studies, including Anello 2009, demonstrate that autism prevalence is four or more times higher in males than in females. Hence, X-chromosome genes are hypothesized to be especially important in the etiology of the disease (Smith, Anne, & Flodman, 2009).
In this study, the tertiary structures of various NLGN3 isoforms are predicted using threading modeling. These structures are then equilibrated and optimized by applying atomistic in order to gain deeper understanding of the interaction between neurexin 1-beta and various isoforms of NLGN3 as well as create a fine protein structures for NLGN3 isoforms and NRX1β
Results:
1. I-TASSER is a comprehensive web-server tool to generate threedimensional structures. Rosetta and NAMD software are able to create docking NLGN3 and NRX1β complex and then refine those structures.
2. Rosetta result show the unfavorable binding between NLGN3 and NRX1β when missing exons, especially exon 7. NAMD energy output shows that wild type V1 and V2 have more capacity of predicting the correct conformation.